|
Peano
|
|
Peano
|
Peano's postprocessing scripts use Python 3. This means you need a Paraview version with Python 3 support (oldish Paraview installations only support Python 2.7, and some Paraview downloads don't come along with Python support at al).
You can display Peano's patch files directly within Paraview. Again, there are different ways. For all of them, Peano's Python environment has to be available within Paraview:
export PYTHONPATH=Peano-dir/python
Please consult the remarks on third party software on FAQs and Troubleshooting (User Perspective) for further troubleshooting.
You can either manipulate all data directly within Paraview or use Paraview's Python terminal to convert data as a batch from Peano's native format into native Paraview data. I recommend the direct Paraview terminal route to interactively explore data. The actual data processing (of time-dependent data) however can be very slow. Once I know what information I need, I thus usually switch to Paraview's Python interpreter to convert all data into Paraview's native file formats in one rush.
For the batch mode, open the terminal pvpython. For the interactive GUI mode, open Paraview and activate the Python Shell by clicking the check box in the View menu.
Both visualisation routes first require you to load Peano's Python tools - either within the pvpython terminal or the Paraview Python window:
import peano4.visualisation
Peano provides a Python class peano4.visualise.Visualiser which allows you to navigate through the whole data.
The snippet displays the first dataset (time step) from a data dump. Via
you can, for example, select the 11th data set and display this one.
Running through data sets (time steps) manually is cumbersome in many cases. We have not yet managed to integrate our Python-based, bespoke postprocessing into Paraview's video replay features, and we also experience that it is relatively slow. In such a case, it is more convenient to convert all data in one rush into Paraview's native, binary file format vtu:
Once this routine terminates, you'll find a file with the extension .pvd in your directory which you can open and play as video. Obviously, this conversion can be triggered in Paraview's Python interpreter which is typically faster than Paraview's built-in terminal.
You can trigger the Python postprocessing routines while the simulation is running.
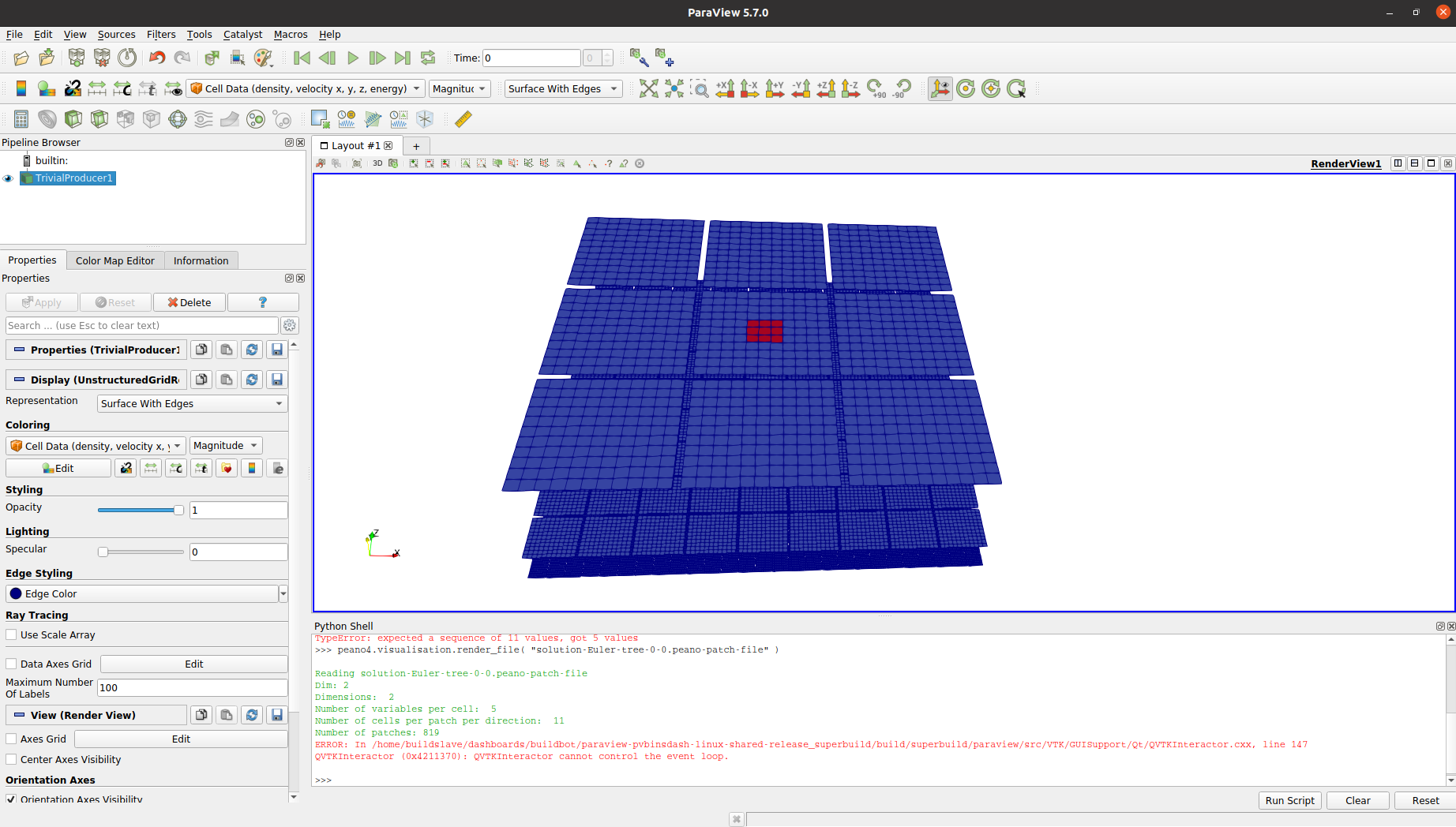
Load the file you are interested in via
This is only one snapshot file, i.e. one file written by one thread in one step. We can display the file with
If we write large parallel files or time series, Peano will typically create one file like solution-Euler.peano-patch-file which links to all the files such as solution-Euler-tree-0-0.peano-patch-file. The meta file (with the links) is plain text, so you can study it via a text editor. Instead of picking a particular file, you can display a particular data set (snapshot) from the meta file:
Peano's snapshots are relatively complex. In practice, you typically want to filter data before you pipe it into Paraview. The most popular filter is the one which eliminates the coarse grid levels: Peano holds all data in multiple resolution. If your code does not exploit this feature, you might not be interested in the coarser resolutions, so removing it makes sense.
There are more filters or you can write your own custom filters. Study the source code for further details.